Transformers are an AI language model that has had huge impact in speech recognition and automated chatbots and has now been pointed out as a revolutionising technique to understand the language of proteins, offering an innovative approach to study and design proteins.
What are transformers?
Transformers are a type of AI model designed to understand and process language. When trained with a large database on a particular language, such as English, the model learns about the words and grammar and can generate complete sentences when prompted. Let’s look at the following example:
I like ___ water
Taking into account the surrounding context, the transformer is capable of inferring a variety of words that fit both in terms of grammar and semantics. In order to identify the missing words, the model leverages knowledge of a large data set of registered sentences. The transformer responds with a set of probable words, with one of the preferable options being the word ‘drinking’, completing the sentence as follows:
I like drinking water
How are transformers being applied to protein research?
How are transformers being applied to protein research?
First, let’s look at what proteins are. Proteins are molecules that perform critical functions in all living beings. For example, haemoglobin is the protein charged with delivering oxygen through our body, and antibodies are proteins used as ammunition to defend ourselves from external agents such as viruses and bacteria.
Proteins consist of one or more strings of amino acids. There are only 20 different amino acids and the different combinations of them have resulted in thousands of functional proteins in humans.
If we consider amino acids as words that constitute proteins, which are the sentences, then we could use transformers to understand the language of proteins. When trained with the billions of protein sequences identified so far across multiple species, a transformer is capable of understanding what sequences of amino acids make sense from a language perspective and can propose new combinations. So for example, for the first 5 amino acids of the human insulin protein:
First 5 amino acids in the human insulin protein
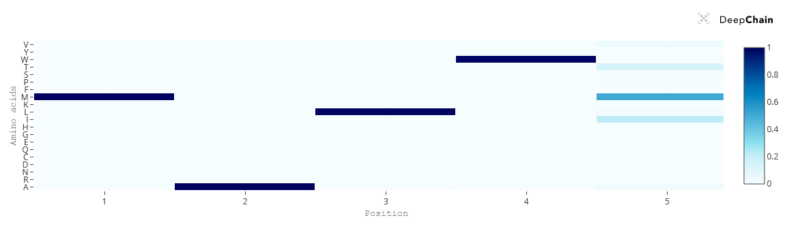
The transformer can tell us that with a 100% probability the first amino acid should be an M (Methionine), which correlates with the fact that most proteins do begin with an M, since it is the signal used by cells to start producing a new protein. The amino acids in positions 2, 3 and 4 also appear to be key; however, position number 5 in this sequence seems to not be so important and the transformer suggests potential variations. Although M is still the most fitting amino acid, the sequence would also accept I, T and V:
Even with such a short sequence, this exercise has told us two things. The first one is that the first 4 positions are recognised as key from a language perspective, and at least for the first amino acid we know this correlates with function. The second one is that position 5 in the sequence seems to be flexible and accept amino acid variations, which indicates that it might not be key for protein structure and/or function.
What’s the powerful impact of transformers in protein research?
Querying a transformer trained in the language of proteins on a particular sequence provides a wealth of information about the protein. As seen in the above example, the transformer can tell you which amino acids might be key and need to be present at the protein of interest from a language perspective. This information is of particular interest when trying to understand amino acid regions that might be essential to protein function or stability.
Results from the transformer can be enriched with insights derived from protein structure research and from comparing how the sequence looks on other species, since evolutionary conserved amino acids tend to associate with key functional areas. But most importantly, the fact that transformers are purely based on language, as explained, is extra helpful in cases where no details are available on the structure of the protein or the species conservation.
Why don’t you try for yourself?
Powered by InstaDeep, whose mission is to accelerate the transition to an AI first world that benefits everyone, DeepChain™ has been launched as a breakthrough innovation tool to accelerate and optimise protein design. It is a cloud-native platform that allows the discovery of new protein designs and validation with molecular dynamics simulations without human interaction, and no ML experience needed.
The DeepChain™ Playground module leverages transformer algorithms and is now accessible for free to analyse your protein sequences of interest and to discover variants and key regions. Create your personalised and secure account and start using AI to accelerate and improve your design process and lead to key discoveries by registering here.
If you would like to learn more about DeepChain™, feel free to send us an email at hello@deepchain.bio !