Protein structure and dynamics are essential for understanding function
Proteins are involved in all of the essential processes in living beings; including metabolite transport, signalling, building tissues, maintaining balance in organisms, and detection and protection from infections. Proteins are produced as unstructured strings of amino acids, which soon adopt the unique 3D structure required for their function.
For many years, scientists have relied on the experimental methods for resolution of protein structures to understand their function in health and disease. Experimental techniques, such as X-ray crystallography, NMR or – recently very successful – cryo-EM, have provided accurate ways to gain insight into the world of protein structure, even though it can not be observed directly. The experimental models, determined by such methods, are deposited in the world-wide Protein Data Bank resources, hosted at RCSB (United States), EMBL/EBI (United Kingdom) and Osaka University (Japan).
The study of these structures leads to a deeper understanding of how proteins work, however, these experimental techniques have limitations in the sense that they only provide a snapshot of the 3D protein structure, leading to static models. We know that proteins are not rigid and in fact their flexibility and conformational changes are key to their function. Kinetics and thermodynamic fluctuations are particularly relevant in proteins involved in transport, such as channels and transporters; molecular recognition, such as antibodies; or chemical reactions such as enzymes. Moreover, to permit for resolving their structures, proteins need to be subjected to often unnatural conditions, which can impede the observed structure too. To overcome these limitations, the scientific community developed computational tools to simulate the behaviour of a protein directly from its 3D structure.
Molecular dynamics simulation is a technique to analyse the physical movement of molecules in a simulated, dynamic, biological environment. It leverages the laws of physics, such as Newton’s equations of motion, to estimate the interaction between the atoms comprising a molecule. This can provide information about conformational changes (kinetics) and thermodynamics at an atomic level, allowing for the study of protein complexes at a much deeper level, with lesser effort, than what could be done experimentally.
Molecular Dynamics made easy
Even though they are tremendously valuable, molecular dynamics simulations require substantial expertise and compute resources. To facilitate the use of such a powerful research tool by all scientists, DeepChain™ has been launched as an AI-powered protein design platform featuring a molecular dynamics simulation module, allowing users to run and visualise simulations in just a few clicks. DeepChain™ combines a simple, yet effective user-friendly interface with world-class NVIDIA A100 GPU supercomputing infrastructure that that returns results in under 24 hours.
As a practical example, we are going to look at the GLUT5 protein, a fructose transporter. Fructose makes up to 50% of table sugar, and upon ingestion, it is transported into the digestive system cells to be used for energy. Cancer cells tend to rely heavily on sugar and an increase on the number of GLUT5 transporters has been linked to several types of cancer. Thus, a deep understanding of the dynamics of this protein is highly interesting for the development of a potential cancer therapeutic (Norimichi et. al, Nature, 2015).
In order to run a molecular dynamics simulation in DeepChain™ we need a PDB file as input. In this case, we selected the bovine GLUT5 structure with PDB ID: 4YB9. The platform returns a 3D representation of the protein structure.

Polymer cartoon and coarse surface representation of the bovine GLUT5 protein’s 3D structure
From here, we can easily launch a molecular dynamics simulation. The platform allows for the personalisation of three parameters: simulation time, equilibration time and temperature.

Pop up window displaying the three adjustable parameters of DeepChain’s molecular dynamics simulation module
- Simulation time: duration of the simulation in terms of picoseconds, with 1 picosecond being equal to 10−12 of a second
- Equilibration time: prior to the actual simulation, the molecules and surroundings need to undergo a relaxation step that here is expressed as a percentage of the total time of the simulation
- Reference temperature: temperature of the simulated environment in kelvins
Playing with the temperature allows for the exploration of protein behaviour through a range of temperatures, which is particularly interesting when studying thermophilic proteins, or when exploring temperature-accelerated protein kinetics. For the GLUT5 example, we run the simulation with the default parameters with the exception of the simulation time, which we will increase to 10,000 picoseconds corresponding to 10 nanoseconds.
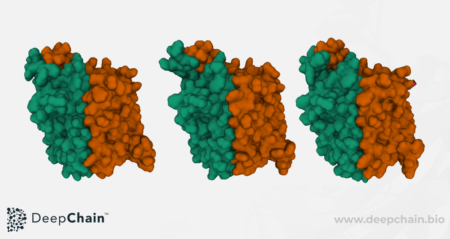
Thanks to DeepChain’s powerful GPUs, results of the molecular dynamics simulation are ready in a few hours (<24h), and are displayed as an interactive visual representation and a graph showing the evolution of the protein energy with time. For the GLUT5 example, the visual representation showcases the two alpha-helix bundles that compose the protein, the C-terminal (orange) and N-terminal (green) as described in (Norimichi et. al, Nature, 2015).

Bovine GLUT5 protein’s molecular dynamics simulation in polymer cartoon and coarse surface representations
The visual representation of the simulation is supported by Mol* offering a wide range of visual personalisation. Multiple representations are supported, including polymer cartoon and coarse surface. The user can also create sections according to molecule type and personalise the colouring. The simulation can also be easily exported as an MP4 video. Check our video below to see how we conduct the simulation described here and download the video file.
In terms of quantitative results, DeepChain™ offers the possibility to examine the potential energy, the kinetic energy, the radius of gyration, the root-mean-square deviation from the starting psoe, and the root-mean-square fluctuation. In this case, we are going to briefly look at the potential energy and the radius of gyration.
In terms of potential energy we would expect to have a steady decline in the energy as the complex stabilises through time. When this does not happen in a neat manner, as it seems to be the case in our example, a longer equilibration time should be used before re-running the simulation. Our example, GLUT5, is a membrane protein and we are expecting high degrees of thermodynamic frustration, since we are running a simulation of the protein in water, as opposed to its natural lipidic, hydrophobic membrane context
As for the radius of gyration (root mean square distance of structure’s atoms to its center of mass); its changes during simulation indicate an undergoing conformational change. If it gets lower, the complex becomes more compact, and more spherical, if it gets notably higher, it means the protein complex has disassembled, partially or completely. Ideally, this metric should decrease slightly, or remain fairly constant after some time, as is the case in our example.

Graphs showing the potential energy and radius of gyration of the bovine GLUT5 dynamics simulation- ran for 10,000 picoseconds, with 10% of equilibration time and at a temperature of 300K
Fancy a live demo?
The DeepChain™ Molecular Dynamics module is now accessible to PRO users to simulate and visualise proteins and complexes of interest quickly and effortlessly.
To learn more about DeepChain™, feel free to send us an email at hello@deepchain.bio!
If you are a computational biologist passionate about AI, please consider joining our team! You can find our job offers for a biology-based position here.