Our bodies are around 15% protein. Our immunity, digestion, how our bodies are structured, regulate themselves and many more functions are the result of cellular processes associated with proteins. Anything that affects protein structure, function, and stability can lead to metabolic disturbance and diseases such as diabetes, infertility, immunity weakness, hormonal disorder. The better we understand their individual functions and how they interplay, the more we strengthen our grasp on how living organisms work and what disturbances might lead to disease. That’s why all of us on the DeepChain™ team are crazy about proteins.
As an AI-powered cloud-native protein design platform, DeepChain™ can be used for several applications related to protein-protein interactions. In this post we’ll use the DeepChain™ AI Designer and particularly its Site Directed Mutagenesis feature, to simulate how a single mutation on a peptide chain can affect its structure and stability – as well as the binding towards a partner protein.
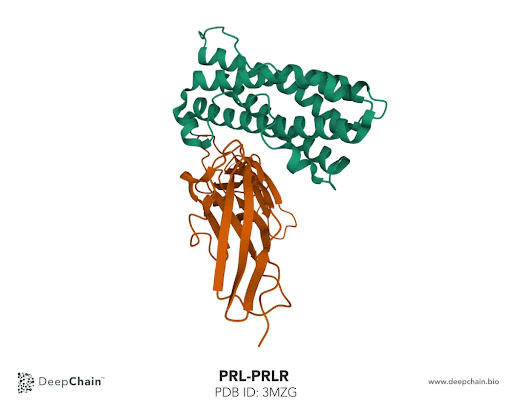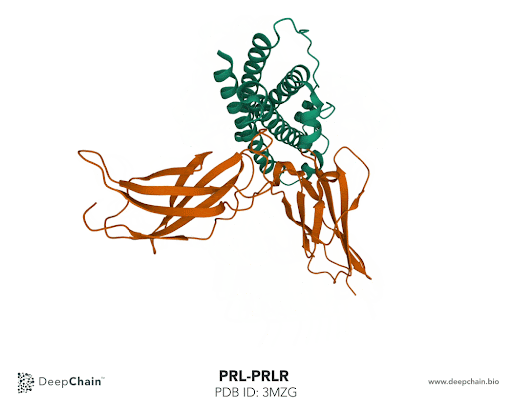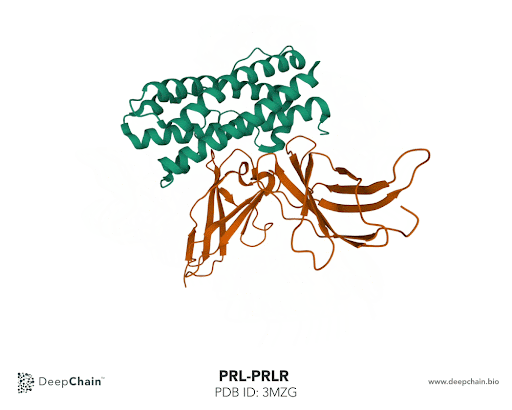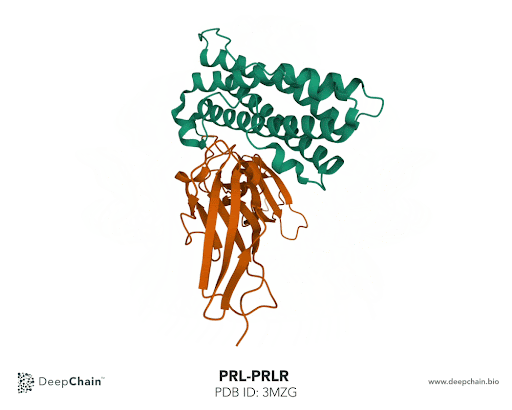
Let’s take the complex formed by prolactin and its receptor as an example. Prolactin is a hormone responsible for lactation, involved in breast development and maternal behavior. A lack of prolactin, or excessive concentration, results in significant metabolic disturbance and may lead to diseases. Using the prolactin and prolactin receptor complex, we’ll show how intuitive the DeepChain™ interface can be and the limitless possibilities it offers.
An easy way to evaluate protein mutations
Proteins are responsible for nearly every task of cellular life: regulation (insulin), transport (hemoglobin), protection (antibodies), structure (collagen) and much more. These macromolecules form through a combination of 20 different smaller building blocks called amino acids. A single change or mutation in an amino acid can affect a protein’s behaviour in various ways, making it unstable, affecting folding, charge and activity among other properties.
In this context, DeepChain™ is a powerful ally for evaluating the consequences of these events.Through its user-friendly interface, DeepChain™ allows us to easily simulate a single mutation (or several) on a protein-protein complex and evaluate its effect on protein stability and binding of the pair.
A single mutation in the prolactin receptor, or PRLR, chain reduces the binding in the PRL-PRLR complex, resulting in high levels of prolactin and leading to infrequent menstrual cycles (oligomenorrhea) and infertility, according to Paul J. Newey et al (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4209110/). Having the information that prolactin doesn’t bind as it should to the mutated receptor, the first line of treatment would be to control the secretion of prolactin. This shows how clarifying the disease mechanism can lead to the appropriate treatment.
We would like to show you how to use the Site-Directed Mutagenesis feature to reproduce this research in silico.
First, sign up to DeepChain™ (https://deepchain.bio/). Once we access workspace home, we can choose one of two ways to design our proteins. The first is we can upload a PDB file, or simply select the option “4-letter PDB ID”. With this option, we just need to write the number of our complex from the Protein Data Bank. PDB is a database for the 3D structural data of large biological molecules such as proteins and nucleic acids. Each file has a four-character PDB identifier. In our case, the identifier for the PRL-PRLR complex is 3MZG.
Once our protein is uploaded, its 3D structure is represented in the bottom with the sequence of the different protein chains at the top. As shown in the GIF below, the two proteins forming the complex have different colors: the green represents the prolactin and the orange the prolactin receptor. We can now select amino acid positions by directly clicking on top of the structure. The position selected will be highlighted in green.
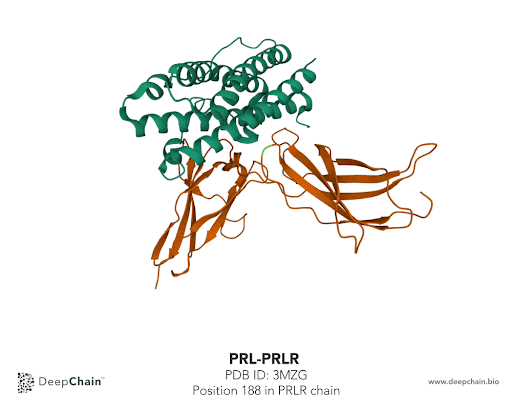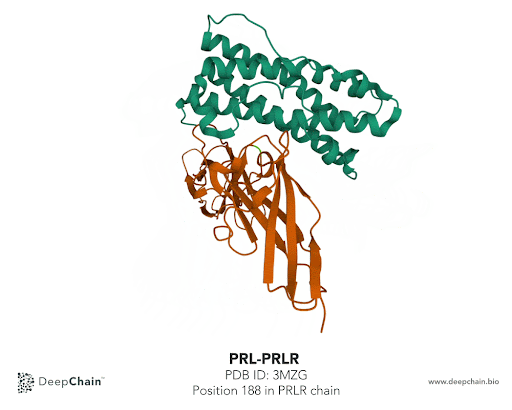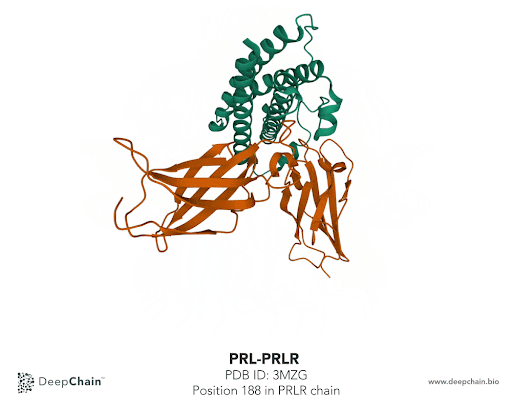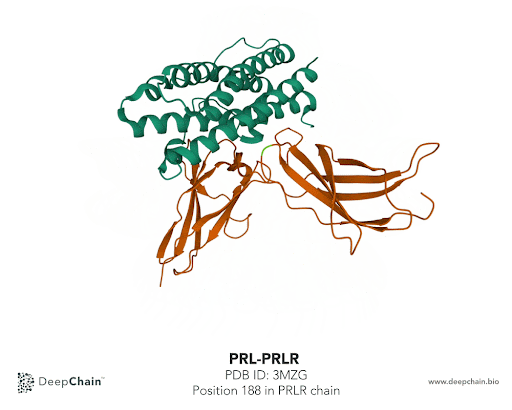
In our case we’ll select the position 188 from the PRLR chain. The native protein contains Histidine (H) residue in this position. According to the paper cited earlier, the mutation affecting this position is H188R. What we’ll do is simply change Histidine to Arginine (R). To do so, we just need to select the position we want to mutate and it will be highlighted in the structure. In the Site-Directed Mutagenesis section will appear H188_, we’ll replace “_” with R and click on “Run Experiment”.
A window pops up allowing us to choose any DeepChain™ App (to learn more check out: https://app.deepchain.bio/hub/apps), in this case we’ll be using the “loglikelihoodApp”. It approximates the likelihood of a protein to appear in nature, and nicely correlates with stability and function. The App calculates the log-likelihood which technically is a negative number, so the closer the result is to 0, the more likely the sequence is to emerge in nature.
On top of the selected App, DeepChain™ uses biophysic and statistical methods, simulations and neural networks trained with billions of protein sequences to score binding, stability and other parameters for the native and mutant protein sequences.
The processing will take a few hours and we will be notified by email once completed. As a result we obtain a table and a graph where we will choose to display binding and stability as parameters.
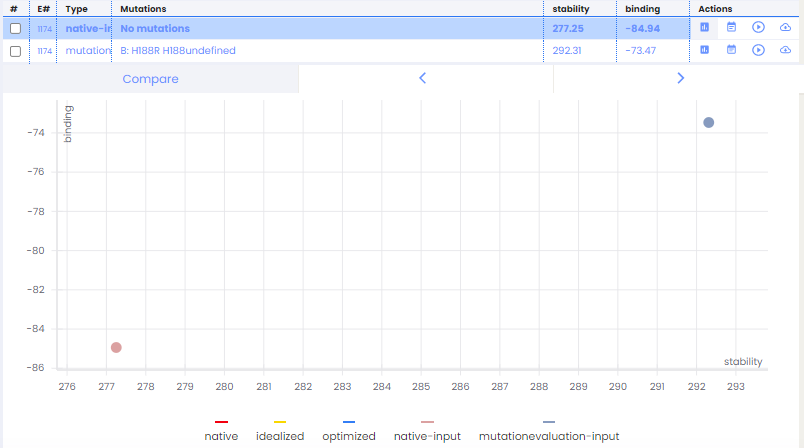
The interpretation of results is based on the fact that the strongest bond and the more stable structure has the lowest free energy. DeepChain™ will calculate this free energy in the native and mutated complex, the lower score is this value for the more stable and/or strongest binding in the complex.
According to the results, we can clearly conclude that the complex containing the mutation (represented in blue/grey) has a lower binding energy and a lower stability than the native complex (represented in pink). Analysing results returned by Site Directed Mutagenesis feature using DeepChain™, we come to the same conclusions reached experimentally.
Experiencing how simple, time and resource-saving is the in silico study of protein- to-protein interaction using DeepChain™, offers a glimpse at the wide range of applications and the impact it can have on drug discovery, disease mechanism elucidation and medical research.
Check out the video below to see yourself how easy it was for us to run the prolactin experiment and for a detailed, step-by-step tutorial.
Why don’t you try for yourself?
Is there a mutation you want to try? Maybe there’s a disease mechanism you want to understand better? We are now offering a week’s FREE trial of our PRO licence, which is specifically thought for researchers and students investigating protein mechanisms.
Sign up here now to try for yourself or feel free to send us an email at hello@deepchain.bio if you want to learn more about how DeepChain™ can accelerate your research today.
If you are a computational biologist passionate about AI, please consider joining our team! You can find our job offers for a biology-based position here.


