When you search for the word “protein”, the first results that appear are related to food, nutrition and diet. Beyond that, proteins are much more than what you eat. It would take pages and pages to list all the functions proteins perform in our organism and all the processes in which they are involved.
Just think of proteins as what allows your body to move. They are the transport network that carries every single molecule to its final destination, they digest and assimilate food and they also defend us from viruses, bacteria and other attacks.
There are so many roles that protein can perform at organism level, and they do much more at cellular scale. In fact, proteins are essential for almost every task, they are the building blocks of living organisms and the fact that they are so numerous reflects their importance.
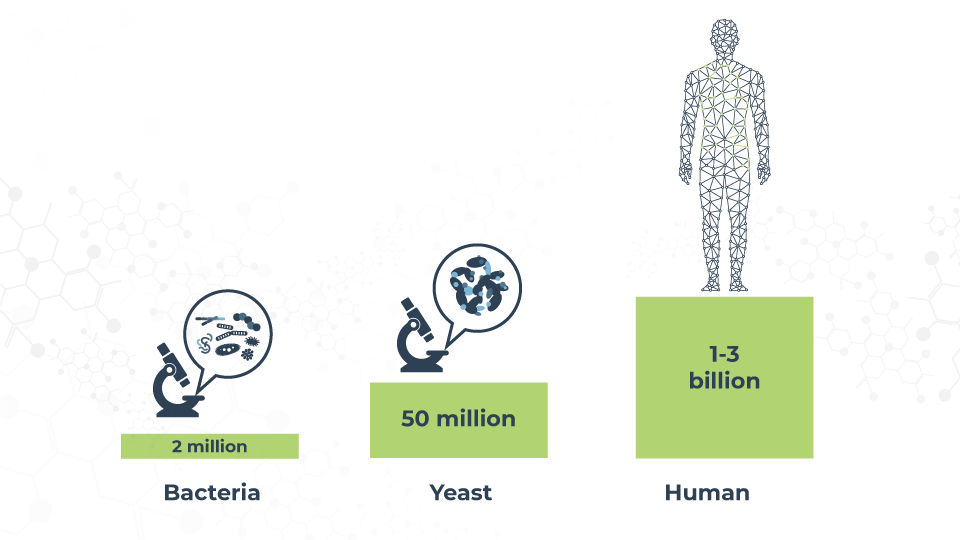
The human body contains somewhere between 80,000 and 400,000 different proteins – so many that we still ignore a lot about them. Considering that each protein can have between 100 and 10,000,000 copies, our knowledge is still just a drop in the ocean.
Due to their abundance, wide distribution and primordial role in cellular function, it’s important to gather as much information as we can about these structures. One of these important pieces of information is how they fold, the space they fill and how they move and interact with their environment. This knowledge is key to understanding main cellular functions, from division to immune response.
Current methods are complex and time-consuming
The process of protein folding is ruled by the laws of physics, and the only way scientists had to demonstrate it relied on complex techniques that come with a set of constraints. The common experimental method for this is x-ray crystallography, which can be done in a few months (if the researcher is lucky, or years if they aren’t). This method relies on changing the protein to solid crystal and capturing every single scatter view to determine the position of each atom.
So far, x-ray crystallography, along with other experimental methods has allowed us to determine the structure of 150,000 proteins, bearing in mind that the total number is estimated to be around 10¹². This means we know the shape of only 0.000015% of what nature offers, and this doesn’t take into account all possible mutations either. Within these near-infinite possibilities lies the protein folding challenge, and the difficulty of creating a method or tool to predict what a protein will look like.
Mastering this prediction ability will open up a wide range of possibilities for the future. This could be the key to discovering cures for diseases such as Alzheimer’s and Parkinson’s, since they are known to be caused by misfolding proteins that disturb brain activity1, 2. And this can be extrapolated to tackle many more diseases by helping us understand their mechanisms and find a way to eradicate them.
The lock and key analogy
Proteins’ mechanism of action has been widely compared to a lock and key process where a lock (enzyme) fits a specific key (substrate) and vice versa. And in these mechanisms lies the importance of shape. This is what allows these complex structures to perform their diverse functions. In a nutshell, shape is function.
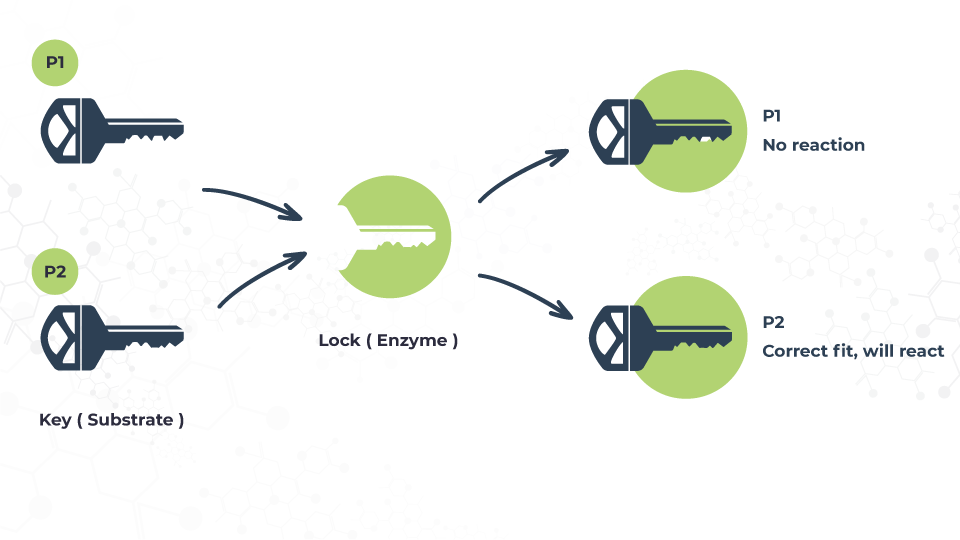
This knowledge can also significantly affect diagnosis methods and effectiveness, just like the example of the lock and key mechanisms. If we know exactly what’s inside the lock, we can design the appropriate key and potentially save time and effort treating the patient.
Relying on our progress in understanding proteins, we have been able to support the process of 3D structure illustration using computational predictions. And today InstaDeep is going a step further in this road making structure prediction available via its DeepChain™ platform.
Thanks to our computational expertise and compute capacities leveraging AI and biology advances, we offer users the power to solve protein folding problems with high accuracy in only a few clicks.
Associating protein structure prediction to the DeepChain™ platform opens the door to anyone interested in protein-protein interaction to run end-to-end in silico experiments. Starting from two or more amino acid sequences, we can fold them together, and with the obtained 3D structure, perform in silico mutagenesis. This process means the vast range of possible outcomes is narrowed down to highlight the best candidate for experimental analysis, saving significant time and resources.
Using the DeepChain structure prediction tool
In the following example, we will demonstrate how DeepChain’s structure prediction tool is as easy as copy-pasting a protein sequence and clicking a few buttons.
Let’s take a look at insulin. This protein hormone is composed of two chains comprising 52 amino acids: GIVEQCCTSICSLYQLENYCN and FVNQHLCGSHLVEALYLVCGERGFFYTPKA.
Step 1: From the homepage select in the left menu the sequence folding icon.
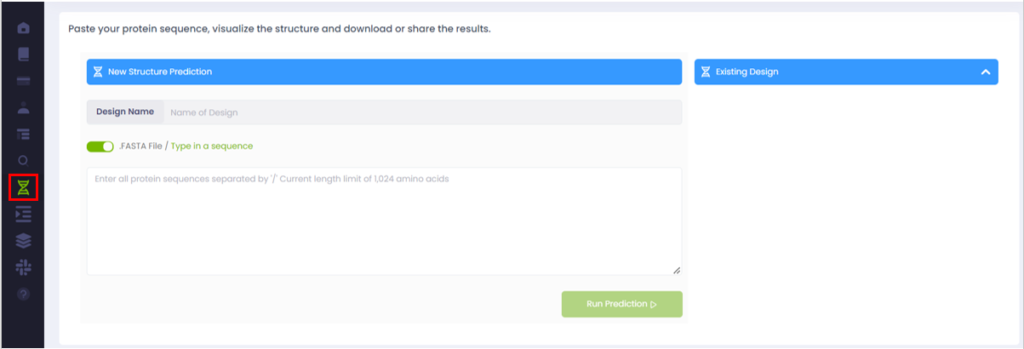
Step 2: Name the sequence and select the input – in our case “type in a sequence” (alternatively, a FASTA file can also be uploaded directly to the platform unchecking the box) – and copy/paste the two chains sequence separated with ‘/’.
Step 3: Click on the “Run Prediction” button. The sequence will be processed by DeepChain™ and appear in the Design tab.
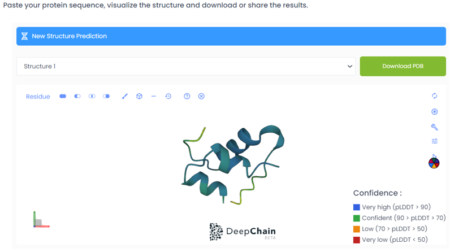
Step 4: When the sequence has been successfully folded, you will be able to select one of the seven results that will be returned by the platform according to the confidence percentage. (“Confidence” is the measure of how reliable or trustworthy the prediction of each amino acid is. In this example we selected Structure 1 and as we can see in the colouring of the folded protein, the prediction is confident to very highly confident.)
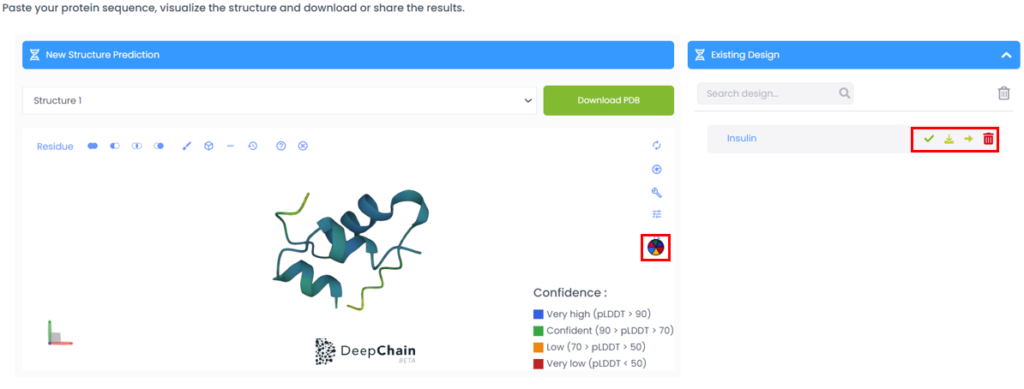
Step 5: The folded structure can be downloaded as a PDB file, or be used directly in DeepChain’s AI Designer. You can see some example experiments in our blog on site directed mutagenesis.
Want to try your own sequences?
Thanks to DeepChain™, you hold all the cards to explore protein sequences from a succession of amino acids to a folded complex and more. If you have a sequence in mind, you can run the steps above and get your own results.
Sign up here now to try for yourself or send us an email at hello@deepchain.bio if you want to learn more about how DeepChain™ can accelerate your research today.
If you are a computational biologist passionate about AI, please consider joining our team! You can find our job offers for a biology-based position here.