Open-source community to stimulate research
AI innovation in biology and healthcare is one of the most exciting intersections in science today thanks to recent breakthroughs such as the use of transformers and the ability to predict protein folding. This flurry of innovation is fueled by the biology community, which regularly produces and shares large datasets, increasing the need to harness AI to maximise insights and make faster and more accurate decisions. However, the required combination of biology expertise, AI technical skills and computing resources often makes this a daunting task.
To address this, DeepChain™ has launched a groundbreaking open-source community where biologists and computer scientists can collaborate on building data- and needs-driven AI tools. We’ve made three repositories publicly available – Bio-Transformers, DeepChain™ Apps and curated Bio-Datasets – to stimulate even more scientific collaboration and provide easy access to AI breakthroughs for everyone.
In this blog, we will dive into the bio-transformers repository, explaining what it is and some of the core applications.
About bio-transformers
Bio-transformers is an open source repository that allows the user to interact with transformer models, pre-trained on millions of protein sequences from the UniRef50, UniRef50S and UniRef90S databases.
Examples of these models are as prot_bert and esm. Upon training, these models understand the language of proteins and are powerful tools for understanding the mutation probability of a protein sequence.
In this example, this probability is often higher in viral proteins than human proteins. Understanding the mutation tolerance of your protein sequence can help to identify potential stable and functional protein variants. This means that probabilities coming from the transformer can be used to predict the fitness landscape of a protein.
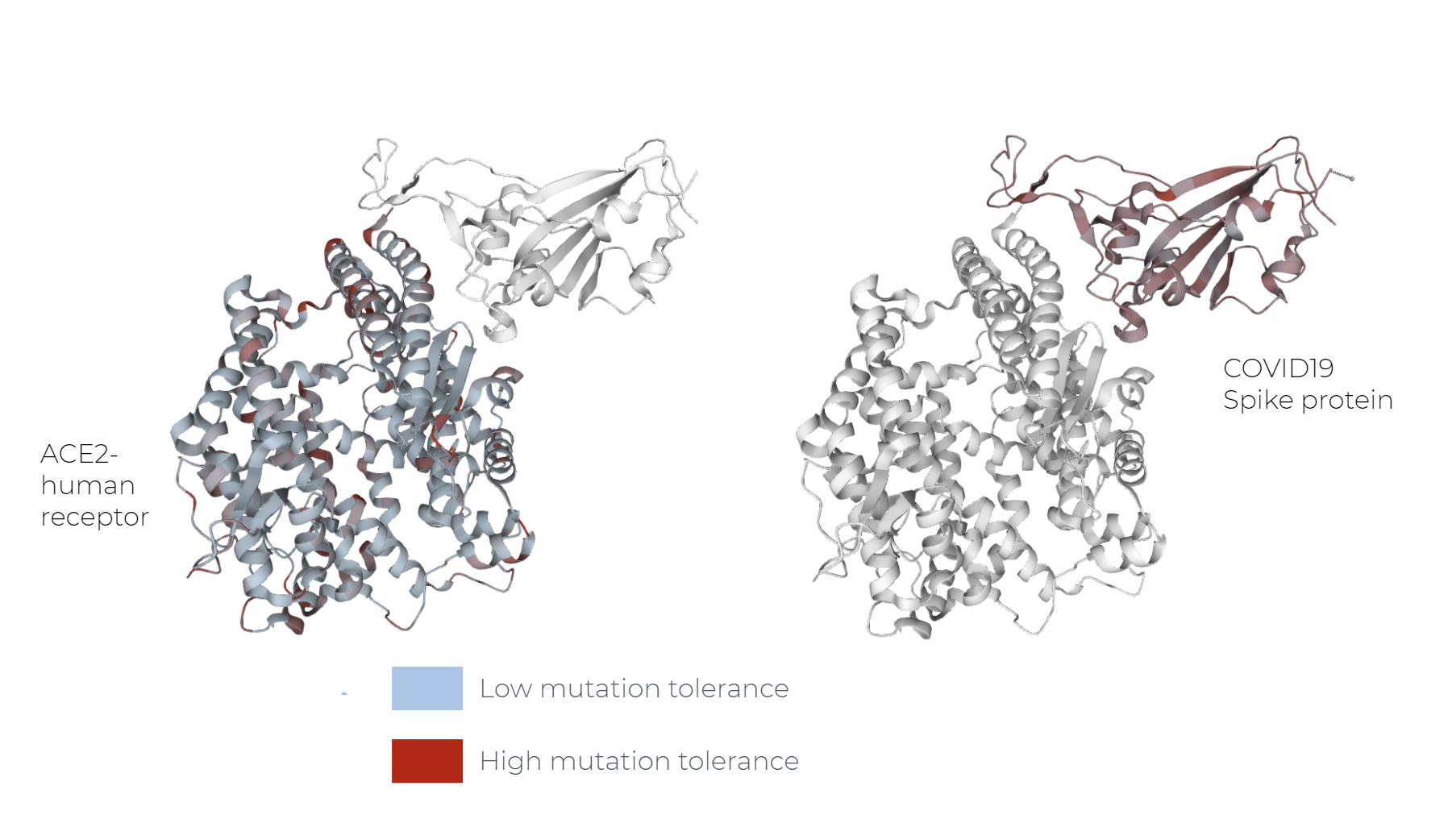
How bio-transformers work: Calculating mutation probabilities step-by-step
Watch this short video to learn how to use the bio-transformer repository, or follow the step-by-step instructions below
Preparation steps:
- Download and install Miniconda from https://docs.conda.io/en/latest/miniconda.html
- Open a Miniconda terminal and create a separate Python environment by typing:
conda create --name bio-transformers python=3.7 -y - Activate the environment with:
conda activate bio-transformers - Proceed to install the bio-transformers repository:
pip install bio-transformers - We recommend moving to Visual Studio Code. Download from https://code.visualstudio.com/.
- In Visual Studio Code, click on File and Create a New File. Then, save it with the extension ‘.py’ to ensure it is recognised as a python file.
- Next step is to install Python. To do this, go to the sidebar on the left and click on extensions. In the search bar, enter ‘Python’ and click on install.
- Once installed, select the Python environment with bio-transformers. To do this, go to the bottom left side of the window and click on the currently in-use Python environment. This should open a menu where you can select the bio-transformers environment. Now everything is ready to start using the transformers.
Using a transformer:
This example will start by computing the embeddings and log-likelihood of four protein sequences.
- The embeddings are a high dimensional representation of the input sequence generated by the neural network. This representation has evolutionary and structural information encoded on it, making it easy to build classifiers or predictors on top of this powerful representation.
- The log-likelihood is an estimation of the probability that a mutated protein will be “natural”. For each amino acid, we estimate the probability of observing this amino acid given the context of the sequence. By multiplying individual probabilities computed for a given amino acid at a particular position within the sequence, we have an estimation of the likelihood, which approximates the stability of the protein.
Compute embeddings and log-likelihood
from biotransformers import BioTransformers # Below we specify the 4 sequences of interest for this example val_seq= ['MAKRVQVVLNETVNKLGRMGQVVEVAPGYARNYLFPRGIAEPATPSALRRVERLQEKERQRLAALKSIAEKQKATLEKLATITIS', 'MAKRVQVVLSEDILSLGKDTVKKQTGGDDVLFGTVTNVDVAEAIESATKKLVDKRDITVPEVHRTGNYKVQVKLHPEVVAEINLEVVSH', 'MAKRVQVVLSQDVYKLGRDGDLVEVAPDVAEAIQVATTQEVDRREITLPEIHKLGFYKVQVKLHADVTAEVEIQVAAL', 'MAKRVQVVLTKNVNKLGKSGDLVEVAILRQVEQRREKERQRLLAERQEAEARKTALQTIGRFVIRKQVGEGRRGITLPEISKTGFYKAQL'] # Selecting the transformer of interest. To see the available ones visit https://github.com/DeepChainBio/bio-transformers bio_trans = BioTransformers("esm1_t6_43M_UR50S", num_gpus=0) embeddings = bio_trans.compute_embeddings(val_seq, pool_mode=('cls','mean')) loglike = bio_trans.compute_loglikelihood(val_seq) print("embeddings: ",embeddings) print("loglikelihood: ", loglike)
Results are provided as an array for both the embeddings and log-likelihood.
- The log-likelihood is the sum of the log-probability of each of the amino acids in the sequence. The closer the result is to 0, the more likely the sequence is to emerge in nature. In the example below, the third sequence with a log-likelihood of -1.894 is the most likely, we can consider it the most favorable.
- Embeddings are a vector representation of the original protein sequence and can be used for training an AI model.

The log-likelihood calculated above is a composition of the probabilities of all amino acids on each given sequence. However, we can also zoom in and calculate the mutation probability for an amino acid of one of our sequences of interest. In this case, we choose the first amino acid of the first sequence.
Compute probabilities for a particular position in a protein sequence
from biotransformers import BioTransformers val_seq = ['MAKRVQVVLNETVNKLGRMGQVVEVAPGYARNYLFPRGIAEPATPSALRRVERLQEKERQRLAALKSIAEKQKATLEKLATITIS', 'MAKRVQVVLSEDILSLGKDTVKKQTGGDDVLFGTVTNVDVAEAIESATKKLVDKRDITVPEVHRTGNYKVQVKLHPEVVAEINLEVVSH', 'MAKRVQVVLSQDVYKLGRDGDLVEVAPDVAEAIQVATTQEVDRREITLPEIHKLGFYKVQVKLHADVTAEVEIQVAAL', 'MAKRVQVVLTKNVNKLGKSGDLVEVAILRQVEQRREKERQRLLAERQEAEARKTALQTIGRFVIRKQVGEGRRGITLPEISKTGFYKAQL'] bio_trans = BioTransformers("esm1_t6_43M_UR50S", num_gpus=0) probabilities = bio_trans.compute_probabilities(val_seq) # We can calculate the probability of the first amminoacid/ token (index 0) of the first sequence: Methionine (M) print("probabilities of M: ",probabilities[0][0])
The output provides the probabilities of each of the 20 different amino acids in the position that we query. In this case, Methionine (M) is the most probable amino acid with a probability over 0.99.

Try for yourself and analyse your protein sequences with innovative language models through our open source community. Start discovering new protein sequences today!
Want to get involved?
Learn more about our Open Source Initiative at www.instadeep.com/open
Read more about DeepChain™ Apps in our post Build powerful AI protein Apps in less than 24 hours with DeepChain™ open-source tools and visit the DeepChain™ GitHub to learn more about the technical aspects of building ML models and engage with our community of experts.
You can also get in touch with us at hello@deepchain.bio. If you are a computational biologist passionate about AI, join our team! You can find our job vacancies here.